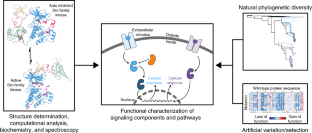
Understanding molecular mechanisms in cell signaling through natural and artificial sequence variation
The functionally tolerated sequence space of proteins can now be explored in an unprecedented way, owing to the expansion of genomic databases and the development of high-throughput methods to interrogate protein function. For signaling proteins, several recent studies have shown how the analysis of sequence variation leverages the available protein-structure information to provide new insights into specificity and allosteric regulation. In this Review, we discuss recent work that illustrates how this emerging approach is providing a deeper understanding of signaling proteins.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
206,07 € per year
only 17,17 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
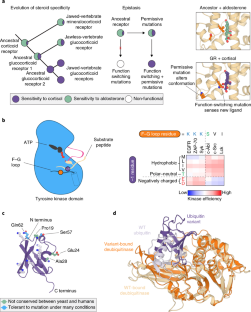
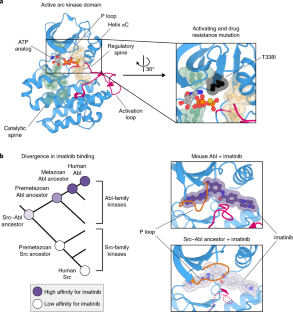
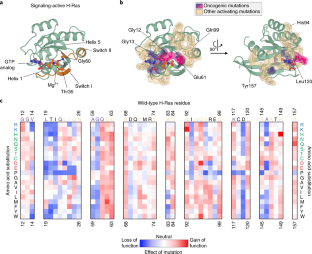

The simplicity of protein sequence-function relationships
Article Open access 11 September 2024

Mapping the energetic and allosteric landscapes of protein binding domains
Article 06 April 2022
Inferring the molecular and phenotypic impact of amino acid variants with MutPred2
Article Open access 20 November 2020
References
- Song, Y. et al. High-resolution comparative modeling with RosettaCM. Structure21, 1735–1742 (2013). ArticleCASPubMedGoogle Scholar
- Webb, B. & Sali, A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics15, 5.6.1–5.6.32 (2014). ArticleGoogle Scholar
- Clapham, D. E. Calcium signaling. Cell131, 1047–1058 (2007). ArticleCASPubMedGoogle Scholar
- Berman, H. M. et al. The cAMP binding domain: an ancient signaling module. Proc. Natl. Acad. Sci. USA102, 45–50 (2005). ArticleCASPubMedGoogle Scholar
- Huang, P., Chandra, V. & Rastinejad, F. Structural overview of the nuclear receptor superfamily: insights into physiology and therapeutics. Annu. Rev. Physiol.72, 247–272 (2010). ArticleCASPubMedPubMed CentralGoogle Scholar
- Kornev, A. P. & Taylor, S. S. Dynamics-driven allostery in protein kinases. Trends Biochem. Sci.40, 628–647 (2015). ArticleCASPubMedPubMed CentralGoogle Scholar
- Shi, Y. Serine/threonine phosphatases: mechanism through structure. Cell139, 468–484 (2009). ArticleCASPubMedGoogle Scholar
- Tonks, N. K. Protein tyrosine phosphatases: from genes, to function, to disease. Nat. Rev. Mol. Cell Biol.7, 833–846 (2006). ArticleCASPubMedGoogle Scholar
- Shah, N. H., Amacher, J. F., Nocka, L. M. & Kuriyan, J. The Src module: an ancient scaffold in the evolution of cytoplasmic tyrosine kinases. Crit. Rev. Biochem. Mol. Biol.53, 535–563 (2018). ArticleCASPubMedPubMed CentralGoogle Scholar
- Pawson, T. & Scott, J. D. Signaling through scaffold, anchoring, and adaptor proteins. Science278, 2075–2080 (1997). ArticleCASPubMedGoogle Scholar
- Lorenz, S., Cantor, A. J., Rape, M. & Kuriyan, J. Macromolecular juggling by ubiquitylation enzymes. BMC Biol.11, 65 (2013). ArticlePubMedPubMed CentralCASGoogle Scholar
- Wittinghofer, A. & Vetter, I. R. Structure-function relationships of the G domain, a canonical switch motif. Annu. Rev. Biochem.80, 943–971 (2011). ArticleCASPubMedGoogle Scholar
- Hilger, D., Masureel, M. & Kobilka, B. K. Structure and dynamics of GPCR signaling complexes. Nat. Struct. Mol. Biol.25, 4–12 (2018). ArticleCASPubMedPubMed CentralGoogle Scholar
- Kuriyan, J. & Eisenberg, D. The origin of protein interactions and allostery in colocalization. Nature450, 983–990 (2007). ArticleCASPubMedGoogle Scholar
- O’Rourke, L. & Ladbury, J. E. Specificity is complex and time consuming: mutual exclusivity in tyrosine kinase-mediated signaling. Acc. Chem. Res.36, 410–416 (2003). ArticlePubMedCASGoogle Scholar
- Kapp, O. H. et al. Alignment of 700 globin sequences: extent of amino acid substitution and its correlation with variation in volume. Protein Sci.4, 2179–2190 (1995). ArticleCASPubMedPubMed CentralGoogle Scholar
- de Juan, D., Pazos, F. & Valencia, A. Emerging methods in protein co-evolution. Nat. Rev. Genet.14, 249–261 (2013). ArticlePubMedCASGoogle Scholar
- Marks, D. S. et al. Protein 3D structure computed from evolutionary sequence variation. PLoS One6, e28766 (2011). ArticleCASPubMedPubMed CentralGoogle Scholar
- Morcos, F. et al. Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc. Natl. Acad. Sci. USA108, E1293–E1301 (2011). ArticleCASPubMedPubMed CentralGoogle Scholar
- Lockless, S. W. & Ranganathan, R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science286, 295–299 (1999). ArticleCASPubMedGoogle Scholar
- Halabi, N., Rivoire, O., Leibler, S. & Ranganathan, R. Protein sectors: evolutionary units of three-dimensional structure. Cell138, 774–786 (2009). ArticleCASPubMedPubMed CentralGoogle Scholar
- Süel, G. M., Lockless, S. W., Wall, M. A. & Ranganathan, R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat. Struct. Biol.10, 59–69 (2003). ArticlePubMedCASGoogle Scholar
- Teşileanu, T., Colwell, L. J. & Leibler, S. Protein sectors: statistical coupling analysis versus conservation. PLOS Comput. Biol.11, e1004091 (2015). ArticlePubMedPubMed CentralCASGoogle Scholar
- Pauling, L., Zuckerkandl, E., Henriksen, T. & Lövstad, R. Chemical paleogenetics:. molecular ‘restoration studies’ of extinct forms of life. Acta Chem. Scand.17 (Suppl.), 9–16 (1963). ArticleGoogle Scholar
- Malcolm, B. A., Wilson, K. P., Matthews, B. W., Kirsch, J. F. & Wilson, A. C. Ancestral lysozymes reconstructed, neutrality tested, and thermostability linked to hydrocarbon packing. Nature345, 86–89 (1990). ArticleCASPubMedGoogle Scholar
- Stackhouse, J., Presnell, S. R., McGeehan, G. M., Nambiar, K. P. & Benner, S. A. The ribonuclease from an extinct bovid ruminant. FEBS Lett.262, 104–106 (1990). ArticleCASPubMedGoogle Scholar
- Bowie, J. U., Reidhaar-Olson, J. F., Lim, W. A. & Sauer, R. T. Deciphering the message in protein sequences: tolerance to amino acid substitutions. Science247, 1306–1310 (1990). ArticleCASPubMedGoogle Scholar
- Reidhaar-Olson, J. F. & Sauer, R. T. Combinatorial cassette mutagenesis as a probe of the informational content of protein sequences. Science241, 53–57 (1988). ArticleCASPubMedGoogle Scholar
- Lim, W. A. & Sauer, R. T. Alternative packing arrangements in the hydrophobic core of lambda repressor. Nature339, 31–36 (1989). ArticleCASPubMedGoogle Scholar
- Azam, M., Latek, R. R. & Daley, G. Q. Mechanisms of autoinhibition and STI-571/imatinib resistance revealed by mutagenesis of BCR-ABL. Cell112, 831–843 (2003). ArticleCASPubMedGoogle Scholar
- Nagar, B. et al. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell112, 859–871 (2003). ArticleCASPubMedGoogle Scholar
- Hantschel, O. et al. A myristoyl/phosphotyrosine switch regulates c-Abl. Cell112, 845–857 (2003). ArticleCASPubMedGoogle Scholar
- Lee, B. J. & Shah, N. P. Identification and characterization of activating ABL1 1b kinase mutations: impact on sensitivity to ATP-competitive and allosteric ABL1 inhibitors. Leukemia31, 1096–1107 (2017). ArticleCASPubMedGoogle Scholar
- Mukherjee, S. et al. Genomes OnLine Database (GOLD) v.6: data updates and feature enhancements. Nucleic Acids Res.45(D1), D446–D456 (2017). ArticleCASPubMedGoogle Scholar
- Goodwin, S., McPherson, J. D. & McCombie, W. R. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet.17, 333–351 (2016). ArticleCASPubMedGoogle Scholar
- Kosuri, S. & Church, G. M. Large-scale de novo DNA synthesis: technologies and applications. Nat. Methods11, 499–507 (2014). ArticleCASPubMedPubMed CentralGoogle Scholar
- Packer, M. S. & Liu, D. R. Methods for the directed evolution of proteins. Nat. Rev. Genet.16, 379–394 (2015). ArticleCASPubMedGoogle Scholar
- Hochberg, G. K. A. & Thornton, J. W. Reconstructing ancient proteins to understand the causes of structure and function. Annu. Rev. Biophys.46, 247–269 (2017). ArticleCASPubMedPubMed CentralGoogle Scholar
- Fowler, D. M. & Fields, S. Deep mutational scanning: a new style of protein science. Nat. Methods11, 801–807 (2014). ArticleCASPubMedPubMed CentralGoogle Scholar
- Rojas, A. M., Fuentes, G., Rausell, A. & Valencia, A. The Ras protein superfamily: evolutionary tree and role of conserved amino acids. J. Cell Biol.196, 189–201 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Manning, G., Plowman, G. D., Hunter, T. & Sudarsanam, S. Evolution of protein kinase signaling from yeast to man. Trends Biochem. Sci.27, 514–520 (2002). ArticleCASPubMedGoogle Scholar
- Laudet, V., Hänni, C., Coll, J., Catzeflis, F. & Stéhelin, D. Evolution of the nuclear receptor gene superfamily. EMBO J.11, 1003–1013 (1992). ArticleCASPubMedPubMed CentralGoogle Scholar
- Fredriksson, R., Lagerström, M. C., Lundin, L.-G. & Schiöth, H. B. The G-protein-coupled receptors in the human genome form five main families: phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol.63, 1256–1272 (2003). ArticleCASPubMedGoogle Scholar
- Ortlund, E. A., Bridgham, J. T., Redinbo, M. R. & Thornton, J. W. Crystal structure of an ancient protein: evolution by conformational epistasis. Science317, 1544–1548 (2007). ArticleCASPubMedPubMed CentralGoogle Scholar
- Bridgham, J. T., Ortlund, E. A. & Thornton, J. W. An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature461, 515–519 (2009). ArticleCASPubMedPubMed CentralGoogle Scholar
- Harms, M. J. & Thornton, J. W. Historical contingency and its biophysical basis in glucocorticoid receptor evolution. Nature512, 203–207 (2014). ArticleCASPubMedPubMed CentralGoogle Scholar
- McKeown, A. N. et al. Evolution of DNA specificity in a transcription factor family produced a new gene regulatory module. Cell159, 58–68 (2014). ArticleCASPubMedPubMed CentralGoogle Scholar
- Anderson, D. W., McKeown, A. N. & Thornton, J. W. Intermolecular epistasis shaped the function and evolution of an ancient transcription factor and its DNA binding sites. eLife4, e07864 (2015). ArticlePubMedPubMed CentralCASGoogle Scholar
- Starr, T. N., Picton, L. K. & Thornton, J. W. Alternative evolutionary histories in the sequence space of an ancient protein. Nature549, 409–413 (2017). ArticleCASPubMedPubMed CentralGoogle Scholar
- Miller, C. J. & Turk, B. E. Homing in: mechanisms of substrate targeting by protein kinases. Trends Biochem. Sci.43, 380–394 (2018). ArticleCASPubMedPubMed CentralGoogle Scholar
- Songyang, Z. et al. Catalytic specificity of protein-tyrosine kinases is critical for selective signalling. Nature373, 536–539 (1995). ArticleCASPubMedGoogle Scholar
- Hutti, J. E. et al. A rapid method for determining protein kinase phosphorylation specificity. Nat. Methods1, 27–29 (2004). ArticleCASPubMedGoogle Scholar
- Shah, N. H. et al. An electrostatic selection mechanism controls sequential kinase signaling downstream of the T cell receptor. eLife5, e20105 (2016). ArticlePubMedPubMed CentralCASGoogle Scholar
- Shah, N. H., Löbel, M., Weiss, A. & Kuriyan, J. Fine-tuning of substrate preferences of the Src-family kinase Lck revealed through a high-throughput specificity screen. eLife7, e35190 (2018). ArticlePubMedPubMed CentralGoogle Scholar
- Cantor, A. J., Shah, N. H. & Kuriyan, J. Deep mutational analysis reveals functional trade-offs in the sequences of EGFR autophosphorylation sites. Proc. Natl. Acad. Sci. USA115, E7303–E7312 (2018). ArticleCASPubMedPubMed CentralGoogle Scholar
- Howard, C. J. et al. Ancestral resurrection reveals evolutionary mechanisms of kinase plasticity. eLife3, e04126 (2014). ArticlePubMed CentralCASGoogle Scholar
- Miller, M. L. et al. Linear motif atlas for phosphorylation-dependent signaling. Sci. Signal.1, ra2 (2008). ArticlePubMedPubMed CentralCASGoogle Scholar
- Creixell, P. et al. Unmasking determinants of specificity in the human kinome. Cell163, 187–201 (2015). ArticleCASPubMedPubMed CentralGoogle Scholar
- Kornev, A. P., Haste, N. M., Taylor, S. S. & Eyck, L. F. T. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc. Natl Acad. Sci. USA103, 17783–17788 (2006). ArticleCASPubMedPubMed CentralGoogle Scholar
- Kornev, A. P., Taylor, S. S. & Ten Eyck, L. F. A helix scaffold for the assembly of active protein kinases. Proc. Natl Acad. Sci. USA105, 14377–14382 (2008). ArticleCASPubMedPubMed CentralGoogle Scholar
- Creixell, P. et al. Kinome-wide decoding of network-attacking mutations rewiring cancer signaling. Cell163, 202–217 (2015). ArticleCASPubMedPubMed CentralGoogle Scholar
- Heredia, J. D. et al. Mapping interaction sites on human chemokine receptors by deep mutational scanning. J. Immunol.200, 3825–3839 (2018). ArticleCASPubMedGoogle Scholar
- Miles, T. F. et al. Viral GPCR US28 can signal in response to chemokine agonists of nearly unlimited structural degeneracy. eLife7, e35850 (2018). ArticlePubMedPubMed CentralGoogle Scholar
- Flock, T. et al. Selectivity determinants of GPCR-G-protein binding. Nature545, 317–322 (2017). ArticleCASPubMedPubMed CentralGoogle Scholar
- Flock, T. et al. Universal allosteric mechanism for Gα activation by GPCRs. Nature524, 173–179 (2015). ArticleCASPubMedPubMed CentralGoogle Scholar
- Pickart, C. M. & Eddins, M. J. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta1695, 55–72 (2004). ArticleCASPubMedGoogle Scholar
- Winget, J. M. & Mayor, T. The diversity of ubiquitin recognition: hot spots and varied specificity. Mol. Cell38, 627–635 (2010). ArticleCASPubMedGoogle Scholar
- Roscoe, B. P., Thayer, K. M., Zeldovich, K. B., Fushman, D. & Bolon, D. N. A. Analyses of the effects of all ubiquitin point mutants on yeast growth rate. J. Mol. Biol.425, 1363–1377 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
- Mavor, D. et al. Determination of ubiquitin fitness landscapes under different chemical stresses in a classroom setting. eLife5, e15802 (2016). ArticlePubMedPubMed CentralGoogle Scholar
- Mavor, D. et al. Extending chemical perturbations of the ubiquitin fitness landscape in a classroom setting reveals new constraints on sequence tolerance. Biol. Open7, bio036103 (2018). ArticlePubMedPubMed CentralGoogle Scholar
- Ernst, A. et al. A strategy for modulation of enzymes in the ubiquitin system. Science339, 590–595 (2013). ArticleCASPubMedGoogle Scholar
- Zhang, W. et al. System-wide modulation of HECT E3 ligases with selective ubiquitin variant probes. Mol. Cell62, 121–136 (2016). ArticleCASPubMedPubMed CentralGoogle Scholar
- Zhang, Y. et al. Conformational stabilization of ubiquitin yields potent and selective inhibitors of USP7. Nat. Chem. Biol.9, 51–58 (2013). ArticleCASPubMedGoogle Scholar
- Tang, C., Iwahara, J. & Clore, G. M. Visualization of transient encounter complexes in protein-protein association. Nature444, 383–386 (2006). ArticleCASPubMedGoogle Scholar
- Hu, J. et al. Kinase regulation by hydrophobic spine assembly in cancer. Mol. Cell. Biol.35, 264–276 (2015). ArticlePubMedCASGoogle Scholar
- Creixell, P. et al. Hierarchical organization endows the kinase domain with regulatory plasticity. Cell Syst.7, 371–383.e4 (2018). ArticleCASPubMedPubMed CentralGoogle Scholar
- Wilson, C. et al. Using ancient protein kinases to unravel a modern cancer drug’s mechanism. Science347, 882–886 (2015). ArticleCASPubMedPubMed CentralGoogle Scholar
- Seeliger, M. A. et al. c-Src binds to the cancer drug imatinib with an inactive Abl/c-Kit conformation and a distributed thermodynamic penalty. Structure15, 299–311 (2007). ArticleCASPubMedGoogle Scholar
- Dar, A. C., Lopez, M. S. & Shokat, K. M. Small molecule recognition of c-Src via the Imatinib-binding conformation. Chem. Biol.15, 1015–1022 (2008). ArticleCASPubMedPubMed CentralGoogle Scholar
- Seeliger, M. A. et al. Equally potent inhibition of c-Src and Abl by compounds that recognize inactive kinase conformations. Cancer Res.69, 2384–2392 (2009). ArticleCASPubMedPubMed CentralGoogle Scholar
- Pitsawong, W. et al. Dynamics of human protein kinase Aurora A linked to drug selectivity. eLife7, e36656 (2018). ArticlePubMedPubMed CentralGoogle Scholar
- Cherfils, J. & Zeghouf, M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev.93, 269–309 (2013). ArticleCASPubMedGoogle Scholar
- Bandaru, P., Kondo, Y. & Kuriyan, J. The interdependent activation of son-of-sevenless and Ras. Cold Spring Harb. Perspect. Med.https://doi.org/10.1101/cshperspect.a031534 (2018). ArticleGoogle Scholar
- Prior, I. A., Lewis, P. D. & Mattos, C. A comprehensive survey of Ras mutations in cancer. Cancer Res.72, 2457–2467 (2012). ArticleCASPubMedPubMed CentralGoogle Scholar
- Bandaru, P. et al. Deconstruction of the Ras switching cycle through saturation mutagenesis. eLife6, e27810 (2017). ArticlePubMedPubMed CentralGoogle Scholar
- Salinas, V. H. & Ranganathan, R. Coevolution-based inference of amino acid interactions underlying protein function. eLife7, e34300 (2018). ArticlePubMedPubMed CentralGoogle Scholar
- Findlay, G. M. et al. Accurate classification of BRCA1 variants with saturation genome editing. Nature562, 217–222 (2018). ArticleCASPubMedPubMed CentralGoogle Scholar
- Pahuja, K. B. et al. Actionable activating oncogenic ERRB2/HER2 transmembrane and juxtamembrane domain mutations. Cancer Cell34, 792–806.e5 (2018). ArticleCASPubMedPubMed CentralGoogle Scholar
- Ma, L. et al. CRISPR-Cas9-mediated saturated mutagenesis screen predicts clinical drug resistance with improved accuracy. Proc. Natl Acad. Sci. USA114, 11751–11756 (2017). ArticleCASPubMedPubMed CentralGoogle Scholar
- Baretić, D. et al. Structures of closed and open conformations of dimeric human ATM. Sci. Adv.3, e1700933 (2017). ArticlePubMedPubMed CentralCASGoogle Scholar
- Bitbol, A.-F., Dwyer, R. S., Colwell, L. J. & Wingreen, N. S. Inferring interaction partners from protein sequences. Proc. Natl Acad. Sci. USA113, 12180–12185 (2016). ArticleCASPubMedPubMed CentralGoogle Scholar
- Gueudré, T., Baldassi, C., Zamparo, M., Weigt, M. & Pagnani, A. Simultaneous identification of specifically interacting paralogs and interprotein contacts by direct coupling analysis. Proc. Natl Acad. Sci. USA113, 12186–12191 (2016). ArticlePubMedCASPubMed CentralGoogle Scholar
- Hopf, T. A. et al. Sequence co-evolution gives 3D contacts and structures of protein complexes. eLife3, e03430 (2014). ArticlePubMed CentralCASGoogle Scholar
- Ovchinnikov, S., Kamisetty, H. & Baker, D. Robust and accurate prediction of residue-residue interactions across protein interfaces using evolutionary information. eLife3, e02030 (2014). ArticlePubMedPubMed CentralGoogle Scholar
- Dror, R. O. et al. Structural basis for nucleotide exchange in heterotrimeric G proteins. Science348, 1361–1365 (2015). ArticleCASPubMedPubMed CentralGoogle Scholar
- Romero, P. A. & Arnold, F. H. Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol.10, 866–876 (2009). ArticleCASPubMedPubMed CentralGoogle Scholar
- Skerker, J. M. et al. Rewiring the specificity of two-component signal transduction systems. Cell133, 1043–1054 (2008). ArticleCASPubMedPubMed CentralGoogle Scholar
- Procaccini, A., Lunt, B., Szurmant, H., Hwa, T. & Weigt, M. Dissecting the specificity of protein-protein interaction in bacterial two-component signaling: orphans and crosstalks. PLoS One6, e19729 (2011). ArticleCASPubMedPubMed CentralGoogle Scholar
- Coyle, S. M., Flores, J. & Lim, W. A. Exploitation of latent allostery enables the evolution of new modes of MAP kinase regulation. Cell154, 875–887 (2013). ArticleCASPubMedPubMed CentralGoogle Scholar
Acknowledgements
We thank P. Bandaru and R. Ranganathan for insights and stimulating discussions. N.H.S. is a funded by a Damon Runyon–Dale F. Frey Award for Breakthrough Scientists from the Damon Runyon Cancer Research Foundation. J.K. is funded by NIH grant P01 A1091580.
Author information
Authors and Affiliations
- Department of Chemistry, Columbia University, New York, NY, USA Neel H. Shah
- Department of Molecular and Cell Biology, University of California, Berkeley, Berkeley, CA, USA John Kuriyan
- Department of Chemistry, University of California, Berkeley, Berkeley, CA, USA John Kuriyan
- California Institute for Quantitative Biosciences, University of California, Berkeley, Berkeley, CA, USA John Kuriyan
- Howard Hughes Medical Institute, University of California, Berkeley, CA, USA John Kuriyan
- Molecular Biophysics and Integrated Bioimaging Division, Lawrence Berkeley National Laboratory, Berkeley, CA, USA John Kuriyan
- Neel H. Shah








